18:00 Investigation about 00729 ISG ontology
Table of Contents
1 Analysis of GSat performance
The 00729 ontology contains 124 full guarded TGDs and 54 non full guarded TGDs, and 33 unguarded full TGDs are discarded for our experiments. So, it is a small ontology.
Surprisingly, GSat requires more than 5 minutes to compute the saturation of this ontology. The ouputted log shows that GSat derived 467 486 non full TGDs and no full TGD. Moreover, the skolemized version of the saturation requires only 2 seconds to complete the saturation, according the logs. Its outputs contains 24 full TGDs that were not contains in the single headed skolemized input TGDs. It appears that Simple Sat is even faster.
In order to understand how GSat behaves on this ontology, I first printed the derived non full TGDs. Unsurprisingly, I discovered that they have large body and head, for example:
uuid70(GSat_u1) & oboTAO_0001428(GSat_u1) & oboBSPO_0000071(GSat_u1) & oboTAO_0002254(GSat_u1) & oboTAO_0001834(GSat_u1) → exists[GSat_e1,GSat_e2] oboBFO_0000053(GSat_e1,GSat_e2) & oboBFO_0000001(GSat_e1) & obo2BSPO_0000071(GSat_e1) & oboBFO_0000050(GSat_e1,GSat_u1) & oboPATO_0000070(GSat_e2) & obo2TAO_0001834(GSat_e1) & oboTAO_0001421(GSat_e1) & oboBFO_0000002(GSat_e1) & oboBFO_0000004(GSat_e1) & oboBFO_0000052(GSat_e2,GSat_e1) & obo3PATO_0000070(GSat_e1) & obo2TAO_0001428(GSat_e1) & oboBFO_0000051(GSat_u1,GSat_e1) & obo2TAO_0002254(GSat_e1)
Then, I thought that the way the TGDs depends from each other was complex enough to derive many TGDs, when evolving. In order to see these dependencies, I drew the rule dependencies graph of the ontology (see Mugnier, Thomazo (2014). An Introduction to Ontology-Based Query Answering with Existential Rules), which is a graph, where nodes are TGDs and there is a edge from r1 to r2 iff we can apply r2 after deriving some facts from an instance using r1. So, I used the tools Kiabora to compute the rule dependencies graph, on which I built a small graph visualization.
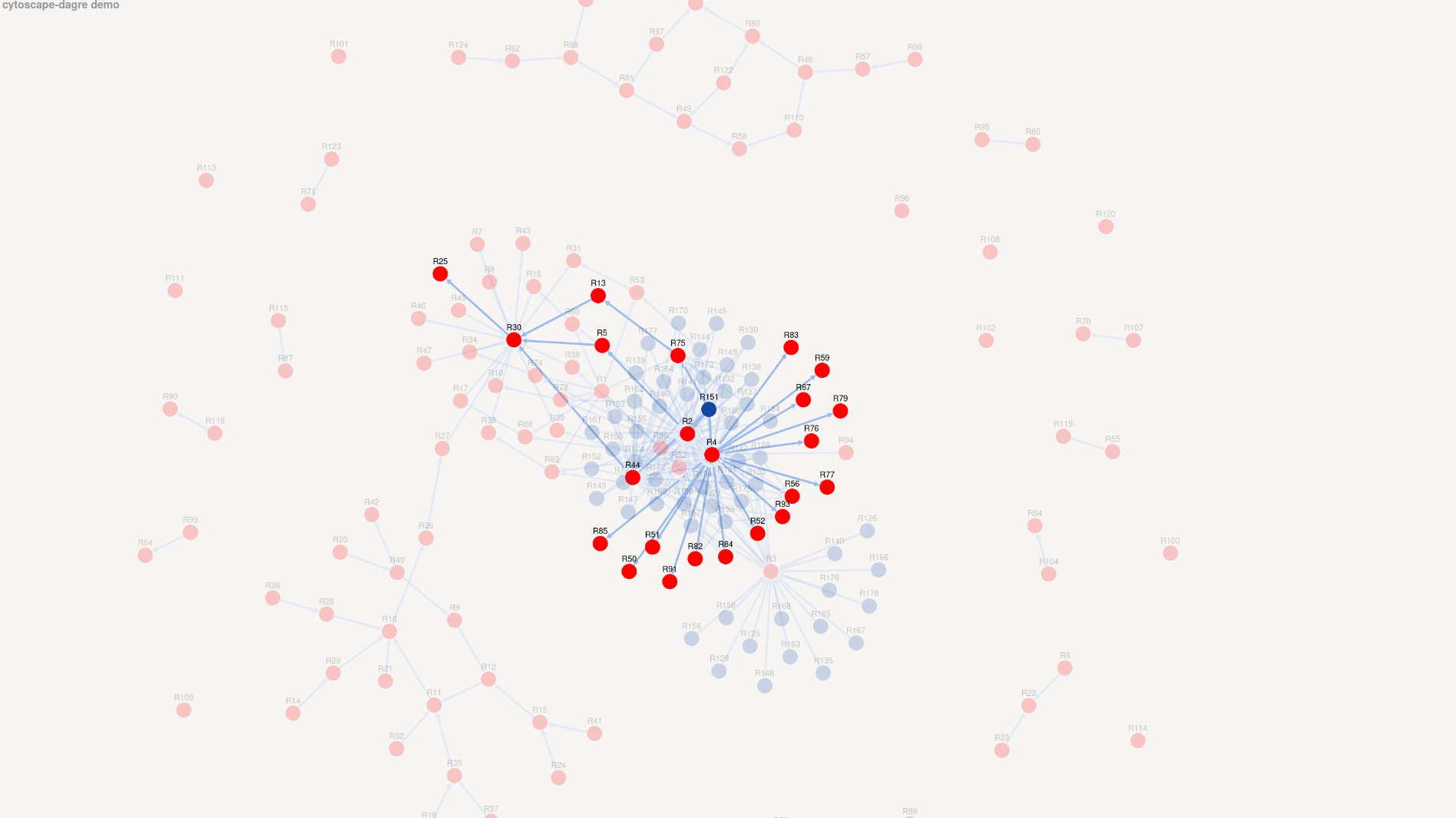
On this picture, you can see the graph, where the red nodes represent the full TGDs and the blue ones the non full TGDs. I discarded the edge which are not useful to understand the behaviour of GSat, i.e. the edges whose target is non full TGDs. We first observe that most of the full TGDs seems not used as an input for evolve, since most of them (red nodes around the group in the centre) are not accessible from a non full TGD (all the blue nodes are in the centre). On the image, you can see all the TGDs accessible from the non full TGD R151, i.e. (need to be proved) all the initial full TGD that may be evolved with R151. I copied these accessible 24 TGDs (see below) into a separate file and ran GSat on them. It tooks almost 1min45s to compile the saturation, which is the set of the initial full TGDs. It did not derived any full TGDs during the execution.
%R2 obo:BFO_0000052(X0, X1) :- obo:BFO_0000053(X1, X0). %R4 obo:BFO_0000050(X0, X1) :- obo:BFO_0000051(X1, X0). %R5 obo:BFO_0000004(X0) :- obo:BFO_0000052(X3, X0). %R13 obo:BFO_0000004(X0) :- obo3:PATO_0000070(X0). %R25 obo:BFO_0000001(X0) :- obo:BFO_0000002(X0). %R30 obo:BFO_0000002(X0) :- obo:BFO_0000004(X0). %R44 obo:BFO_0000004(X0) :- obo:BFO_0000053(X0, X3). %R50 obo2:TAO_0002254(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0002254(X3). %R51 obo2:TAO_0001418(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0001418(X3). %R52 obo2:TAO_0001094(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0001094(X3). %R56 obo2:TAO_0000107(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0000107(X3). %R59 obo2:TAO_0001834(X0) :- obo:TAO_0001834(X3), obo:BFO_0000050(X0, X3). %R67 obo2:BSPO_0000084(X0) :- obo:BFO_0000050(X0, X3), obo:BSPO_0000084(X3). %R75 obo3:PATO_0000070(X0) :- obo:BFO_0000053(X0, X3), obo:PATO_0000070(X3). %R76 obo2:TAO_0001428(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0001428(X3). %R77 obo2:TAO_0001189(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0001189(X3). %R79 obo2:TAO_0000277(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0000277(X3). %R82 obo2:BSPO_0000071(X0) :- obo:BFO_0000050(X0, X3), obo:BSPO_0000071(X3). %R83 obo2:TAO_0001421(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0001421(X3). %R84 obo2:TAO_0001238(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0001238(X3). %R85 obo2:TAO_0000567(X0) :- obo:TAO_0000567(X3), obo:BFO_0000050(X0, X3). %R91 obo2:TAO_0000519(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0000519(X3). %R93 obo2:TAO_0000223(X0) :- obo:BFO_0000050(X0, X3), obo:TAO_0000223(X3). %R151 obo:BFO_0000051(X0, X3), obo:TAO_0001421(X3), obo:BFO_0000053(X3, X4), obo:PATO_0000070(X4) :- uuid:70(X0).
2 Simple example
By analysing the set of these TGDs, we see that there is one non full TGD R151 and two kind of full TGDs the linear ones and the others, that have the following form:
B(x, y), C_i(x) -> D_i(y) for i in 1..n
To simplify, we can remove the linear rule and consider that R151 have the following form:
A(x) -> B(x, y)
Applying Evolve of these TGDs will produce the following TGDs, for I a subset of {1…n}:
R_I : A(x), \wedge_{i \in I} C_i(x) -> B(x, y) \wedge_{i \in I} D_i(y)
Since RI is not subsumed by RJ if I is not equal to J, it means that GSat is creating at least 2n TGDs.
3 Skolemized version
If we simulate the behaviour of the skolemized version of the saturation on the simple example above, we obtain the following input TGDs:
A(x) -> B(x, f(x)) B(x, y), C_i(x) -> D_i(y) for i in 1..n
It is possible to evolve the first non-full TGD with each of the full TGD to obtain the following non-full TGDs:
A(x) -> C_i(x) -> D_i(f(x)) for i in 1..n
This time, the number of derived TGDs is n.