

The UMLS-based reference alignment is known to be incomplete and to contain errors. Turning these reference alignments into an agreed-upon gold standard requires an expert curation process which is very expensive and time consuming due to the size of the input ontologies and alignment sets. Thus, we have opted to move towards a silver standard by harmonising (i.e. voting) the output mappings provided by different matching systems over the relevant ontologies. Furthermore, using this silver standard as reference alignment will also help developers to understand how distant are they outputs wrt the mappings voted by other systems.
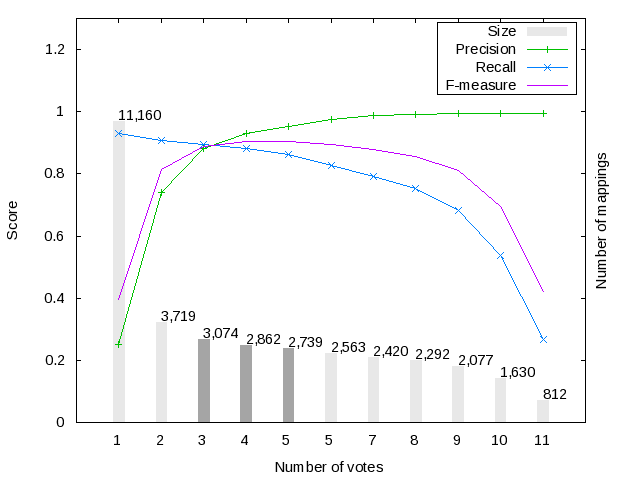
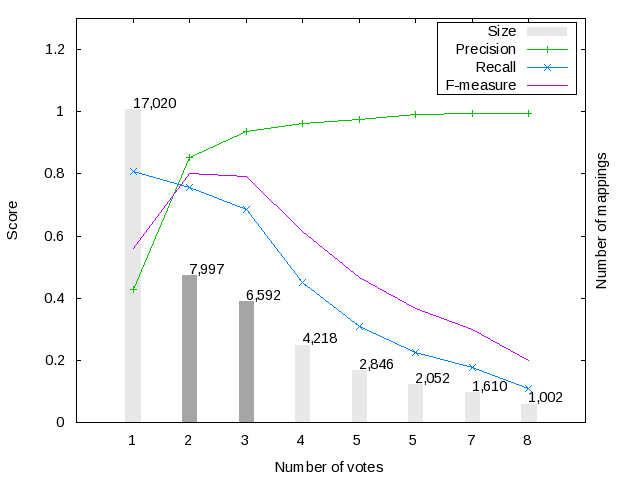
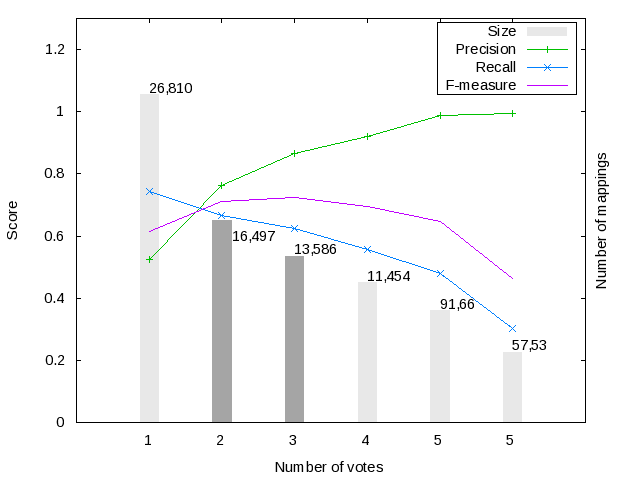
Figure 1 shows the harmonization (i.e. voting) results for the Large BioMed Track matching problems. We have considered only those mappings voted (i.e. included in the output) by at least one system. Mappings have received at most 11 votes from different systems in the FMA-NCI matching problem, while in the FMA-SNOMED and SNOMED-NCI matching problems they got 8 and 5 votes, respectively.
Figure 1 also shows the evolution of F-score, Precision and Recall for the mapping sets with a given number of votes. For example, in the FMA-NCI matching problem, 3,719 mappings have been voted by at least 2 systems and have an F-score of 0.815 wrt the UMLS-based reference alignment. As expected the maximum recall (respectively precision) is reached with the minimum (respectively maximum) number of votes. Note that, in the FMA-NCI problem the recall of the mappings voted by at least 1 system reaches a value of 0.931, while the mappings voted by at least 1 system in the FMA-SNOMED and SMOMED-NCI problems only reach a recall of 0.809 and 0.744, respectively. This clearly shows the difficulty of the FMA-SNOMED and SMOMED-NCI matching problems.
We have selected several silver standard candidates for each matching problem (see dark-grey bars), which have the best trade-off between precision and recall. For the FMA-NCI matching problem we have selected the mappings sets with 3, 4 and 5 votes; while in the FMA-SNOMED and SNOMED-NCI matching problems we have selected mapping sets with 2 and 3 votes.
The voted mapping sets can be downloaded as a zip file (RDF, OWL and TXT formats): voted_mappings_harmo_2012.zip [6.0Mb].
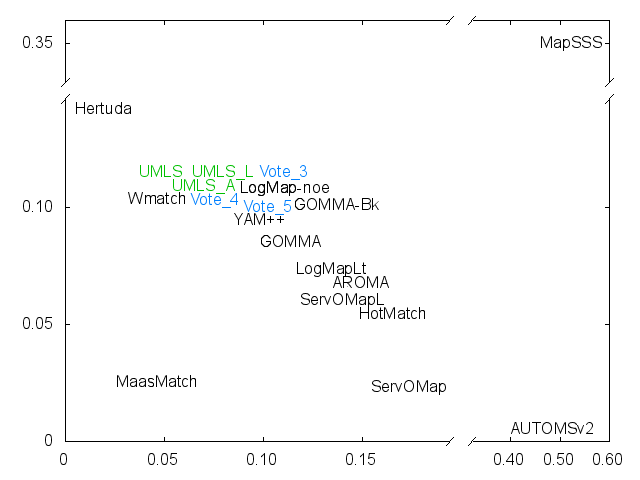
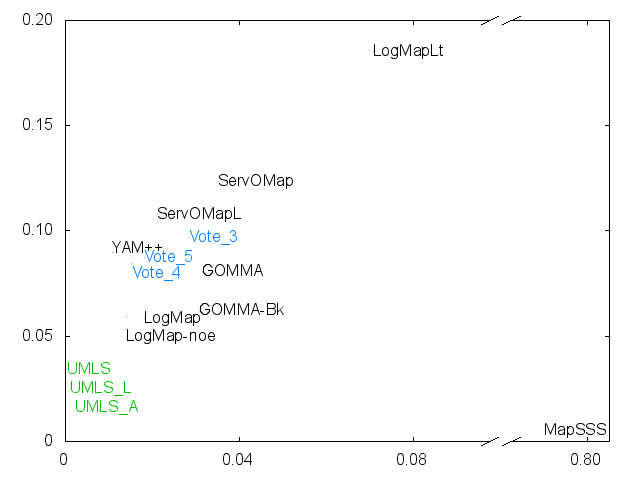
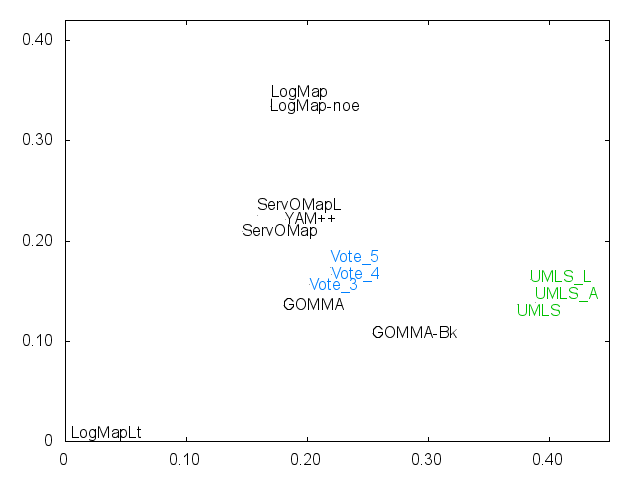
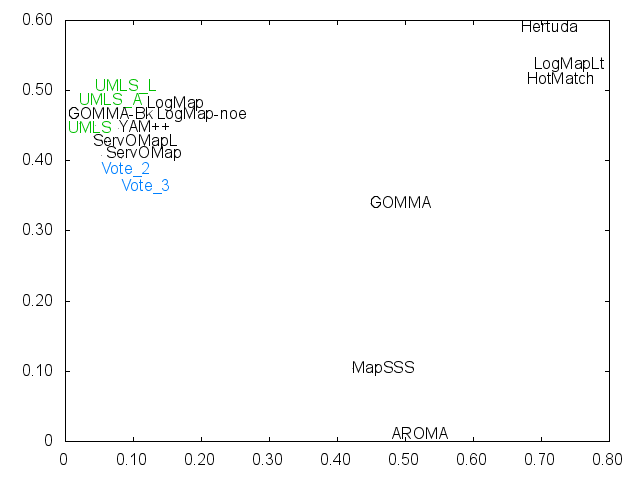
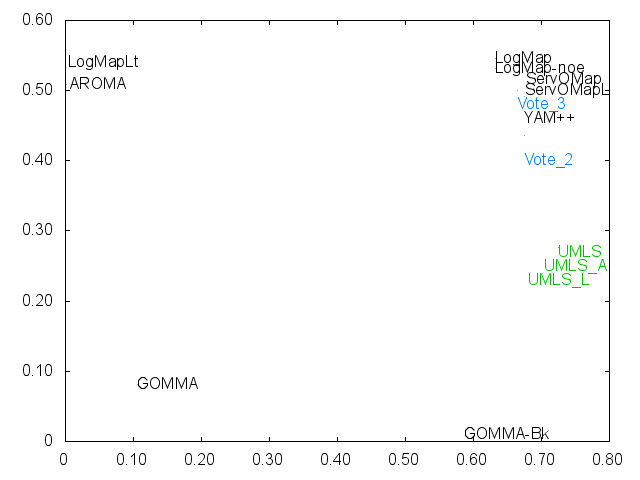
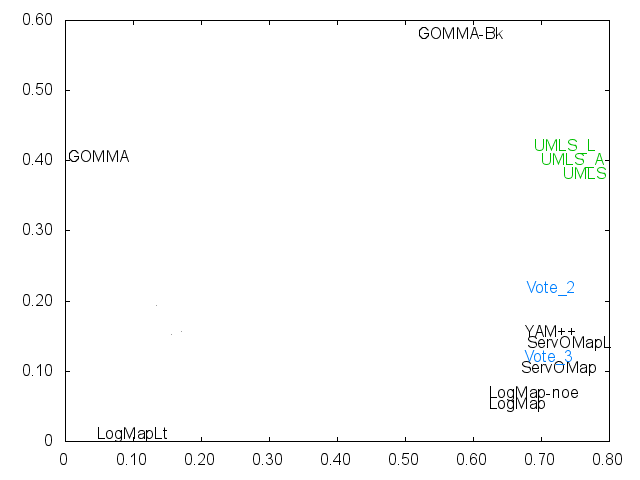
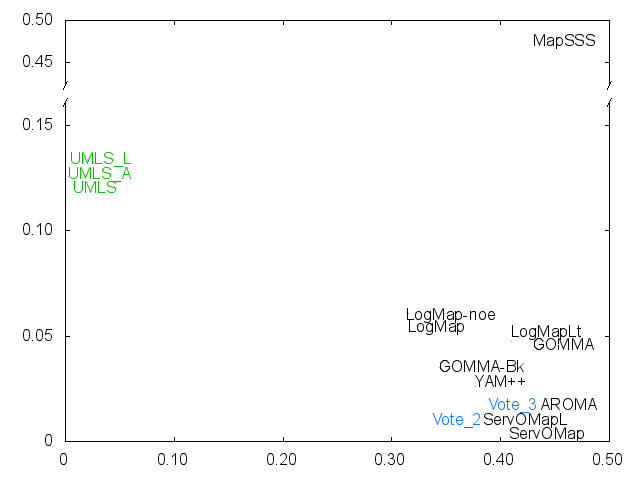
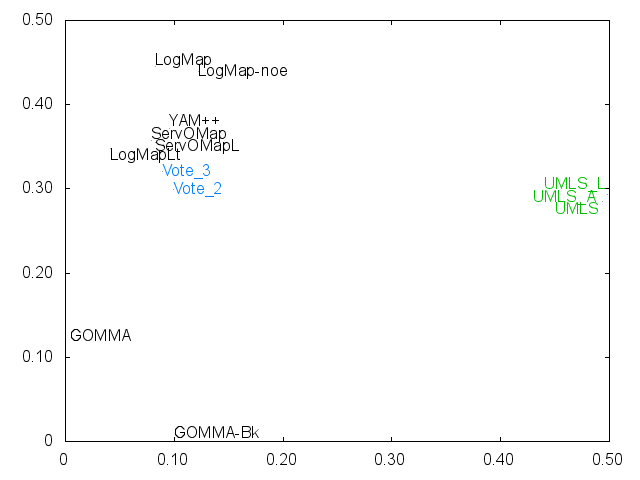
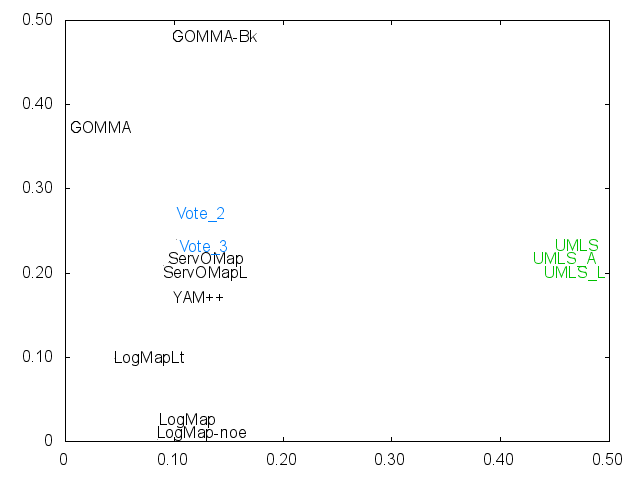
Ernesto Jimenez-Ruiz, Bernardo Cuenca Grau and Ian Horrocks. Is my ontology matching system similar to yours?. The 8th International Workshop on Ontology Matching. [pdf]. October 2013.