Documentation
Below you can find he repository of current options for PyMOSAICS. In order to gain insights into the working of MOSAICS, please see the documentation of the command line version. It might also be helpful to look at the tutorials!
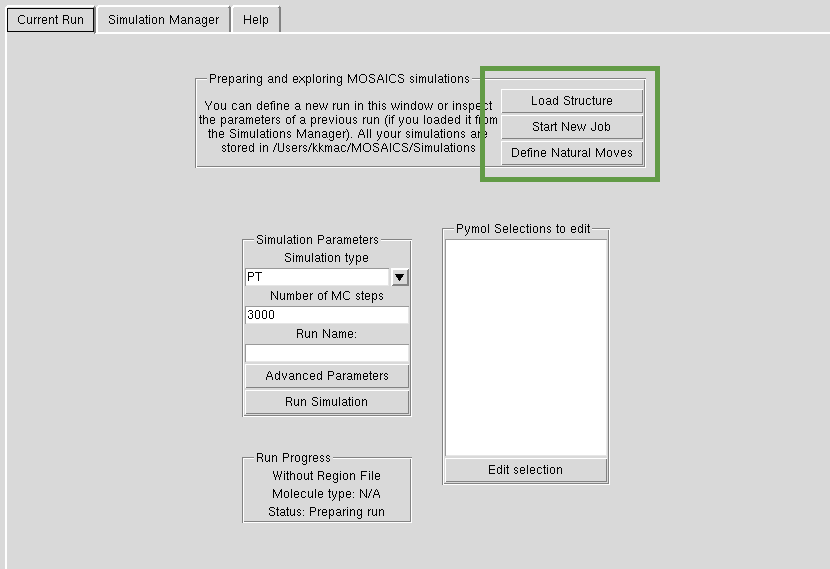
Starting New Jobs
You can start a new job by clicking the 'Start New Job' button as in the screenshot below. If you are currently working on a simulation that had already finished, you will be asked whether to transfer whatever you were doing to a new job. You can load new structures to MOSAICS using the 'Load New Structure' button. Note that MOSAICS needs the structures to be in the precise representation, with all atoms present. In order to fill in the missing atoms we suggest opening the structure first in SwissPdb Viewer which will fill in the missing atoms. For MOSAICS, you should also remove the terminal ends of each chain in the structure. When you have prepared your structure in this way, MOSAICS will try to process it if you are using version >1.7. Just to start with you might want to use an example pdb
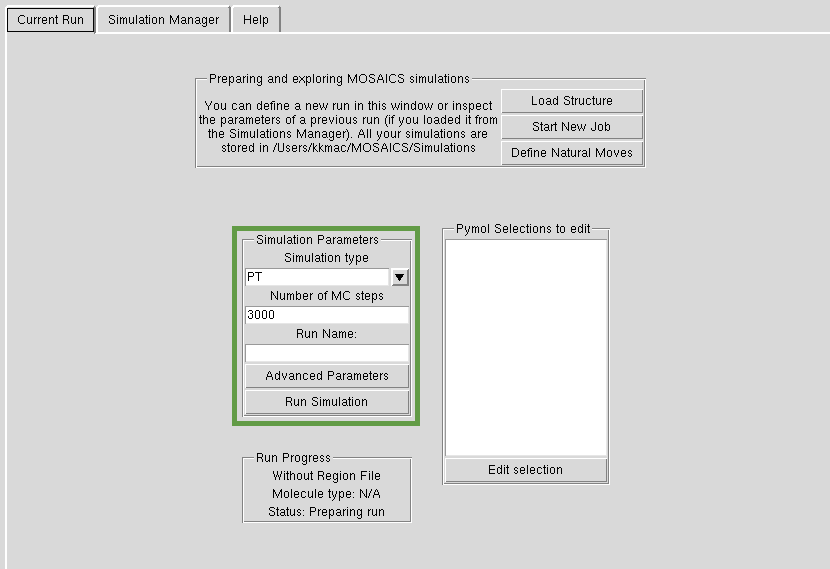
Run Parameters
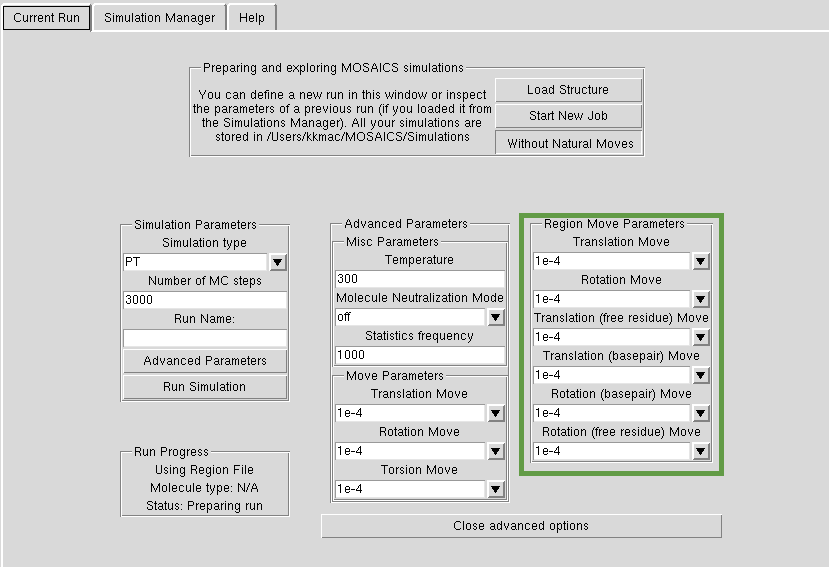
In each MOSAICS simulation you need to choose the sampling methodology such as Parallel Tempering (PT), Normal Modes (NM) etc. In many of our simulations we use PT, if you just want to get started. You also need to tell us how many Monte Carlo simulation steps to perform. If you are just getting the hang of the structure, use something in the order of thousands of steps. For 'proper' simulations, you might to use millions of steps. You give a name to each simulation, which will be the name in the Simulation Manager. You start the job by hitting 'Run Simulation'. NB. for now all the simulations use AMBER bsc0 as the energy function. Please see tutorial to see how this works in practice.
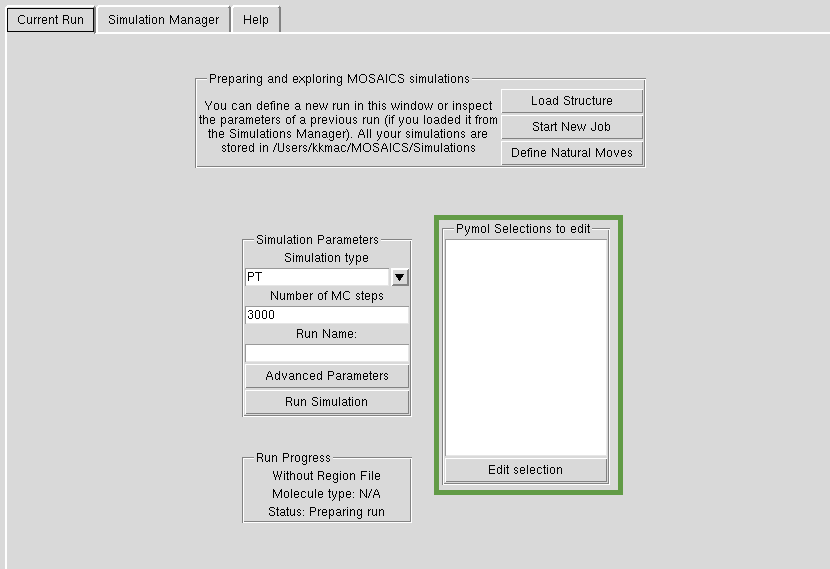
Edit Selections
You can select residues, groups of atoms etc. in Pymos. These selections will be shown in the panel 'Pymol Selections to edit'. You can for instance, select a residue and 'edit' it so add a methylation mark. This is also the tool used to select groups of atoms for natural moves. Please see tutorial to see how this works in practice.
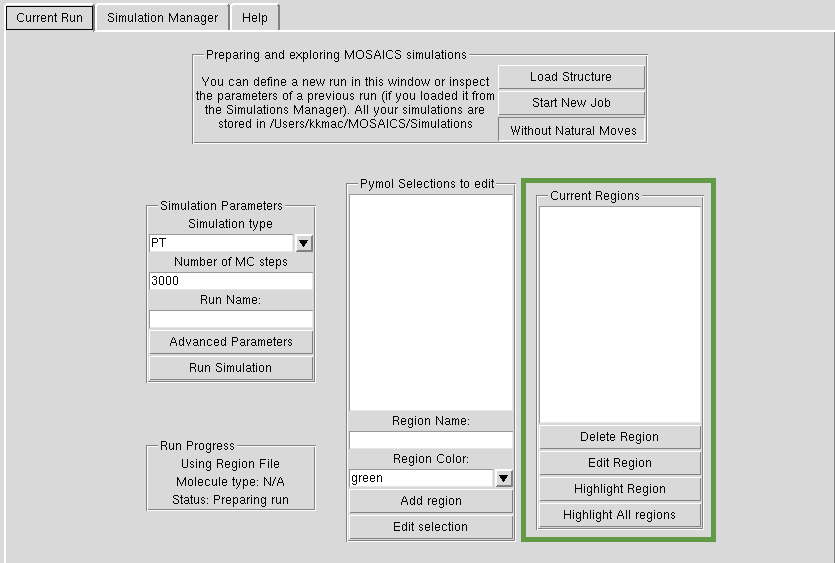
Natural Moves
In order to define Natural Moves, you need to hit the 'Define Natural Moves' button. You can go back on your decision by pressing the same button ('Without Natural Moves'). This will open another panel, 'Current Regions', which holds the Natural Moves that you defined. As shown in tutorials, you select groups of atoms in Pymol, and by pressing 'add region', you create a MOSAICS region. Please see tutorial to see how this works in practice.
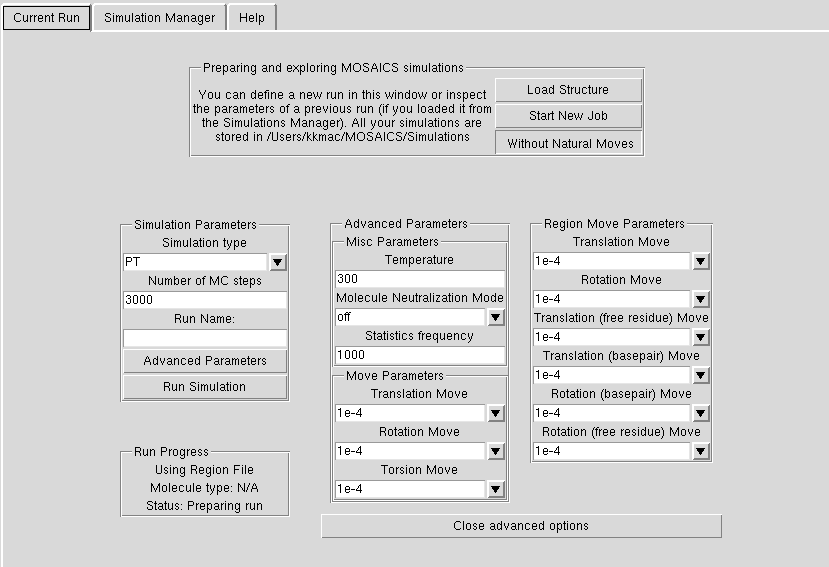
Advanced Options
If you hit the 'Advanced Options' button, you will see some more options that you can define (see below for more detailed descriptions).
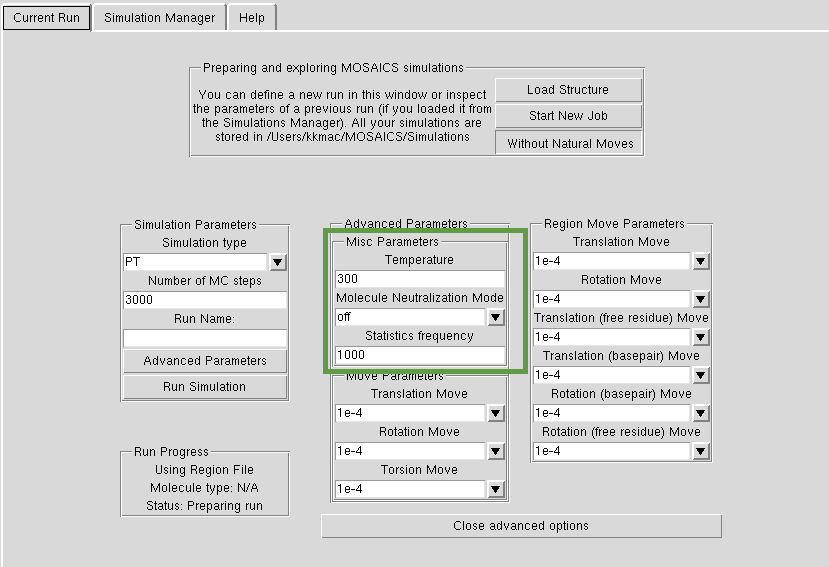
Advanced Options Misc
You can define the 'Temperature' of the simulation (Kelvin). For our simulations we use the value of 300. If you are just getting the hang of your structure, use a much higher temperature (30000) as in tutorials. Statistics frequency describes how often we give you a frame for a trajectory out of the number of Monte Carlo steps you have defined. Thus if you have statistics frequency=200 and MC steps=1000, your simulation will yield 1000/200=5 frames in the final trajectory. Neutralization refers to the method of 'salt addition' as in MD -- for nucleic acids we would normally use the 'nucl' option.
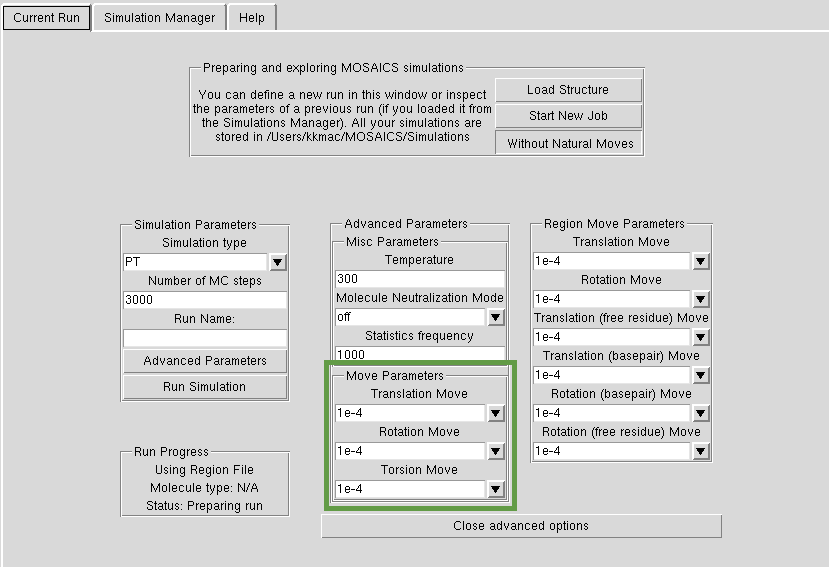
Advanced Options General Moves
Atoms can be given three types of moves: torsional, rotational and translational. The lower the value, the smaller the attempted step. If you put the temerature low and step magnitude high, your sampling will be very inefficient as MOSAICS will try to perform big steps. You should experiment with your structure, with small number of MC steps and different magnitudes of moves, before you start a longer simulation.
Advanced Options Region Moves
If you define Natural Moves, you can give different magnitudes of translational, rotational and torsional values to regions (free residue). Additionally, MOSAICS defines basepair moves, if PyMOSAICS is able to detect basepairing in the regions. Here you define the options each region is created with. In addition you can edit the moves for each region separately, by clicking on the region and hitting 'Edit Region' button in the 'Current Regions' pane.
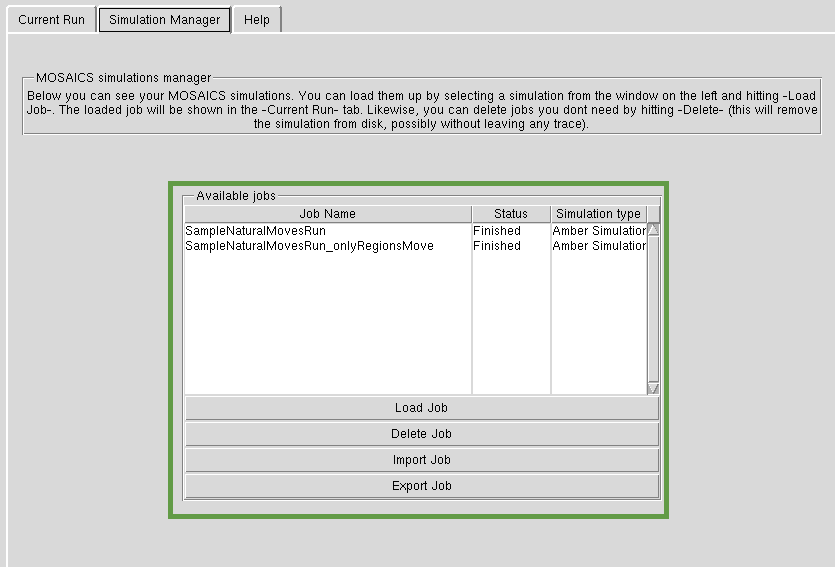
Simulations Manager
Simulations you create are stored in the simulation manager. You can load previous jobs, export, import and delete them. Please see tutorials to see how this works in practice.