Protein Structure Refinement
Hierarchical natural move Monte Carlo has been shown to facilitate the refinement of molecular conformations directly against projections of single particles measured by electron microscopy. By optimizing the orientation of the projection at the same time as the conformation, the method is well-suited to two dimensional class averages from cryoelectron microscopy. The approach has been applied to refine conformations of Methonococcus maripaludis chaperonin (Mm-cpn), a molecular complex with 16 subunits, which form two chambers facilitating the folding of proteins. Below we show examples of natural moves (degrees of freedom or DoFs) used to perform refinement of the system from the closed state to the open state. Further details on the project can be found in (PNAS 109:9845-9850 (2012)).
| DOFs Definition | First 10,000 steps of refinement | Refined Model (Top View) | |
| At Level 1, each subunit of Mm-cpn is treated as a rigid body. | 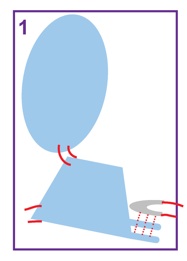 |
 |
|
| At Level 2, equatorial domain and the upper two domains of each subunit are treated as rigid bodies connected by flexible loops. | 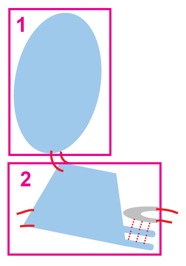 |
 |
|
| At Level 3, even more flexibility is allowed whin each equatorial domain. | 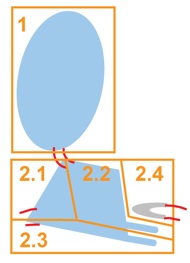 |
 |
|
